(Upload on October 6 2018) [ 日本語 | English ]
Mount Usu / Sarobetsu post-mined peatland
From left: Crater basin in 1986 and 2006. Cottongrass / Daylily
HOME > Lecture catalog / Research summary > Glossary > Chemistry
[ protocol | unit | culture | validation ]
Analytical purposesQuantitative analysis ≈ measurement Lord Kelvin (1824-1907) "If you cannot measure, your knowledge is meager and unsatisfactory."
Qualitative analysis Objects
Inorganic analysis |
Absolute amount of samples
Macro analysis: 0.1-several g Relative amount of objects
Macro determination: 100ppm-100% |
|
|
CHEMFETS (ISFET): Merits = small size (e.g., less than 1 mm² area), low output impedance, in vivo monitoring, small sample volumes, multiple-ion sensor arrays The CHEMIST is essentially a conventional insulated-gate field effect transistor that has its metallic gate contact replaced by a chemically sensitive coating and a reference electrode. Ion-controlled diodesThis device is a combination of a p-n junction and a metal oxide semiconductor capacitor in which the junction makes contact with the inversion layer of the capacitor. Like CHEMFET, the device can be use as an ion sensor by application of a suitable ion-selective coating to the gate surface.Schottky diodesIn its simplest form Schottky diode consists of a small area of metal contact with a semiconductor. This contact exhibits a rectifying behavior and a characteristic nonlinear current-voltage dependence. |
ChemresistorsIn the case of organic semiconductors the use of such compounds as vapor sensors has been hindered by the rather high resistivities of the material (e.g., 108-1015 ohm·cm).Thin-film tin oxide gas sensorsTin oxide and other metal oxide semiconductor are sensitive to low concentration of vapors. In operation a sintered block of the semiconductor is heated to several hundred degrees centigrade and its electrical conductivity is monitored.MicrodielectrometerThe device, presently manufactured by Micromet, consists of a planer interdigital microelectrode array, one side of which is attached as a floating gate to an onchip FET charge amplifier. The other side of the microelectrode array is driven with a sinusoidal voltage. By measuring the amplitude and phase differences of the signal applied to the driven gate and the signal produced by floating gate, it is possible to determine the complex impedance (i.e., the resistance and capacitance) of the medium in contact with the electrode. The integration of the FET amplifier and microelectrode permits extremely weak currents to be measured easily. |
Table 1. Comparison of properties of carbon and reversed-phase silica packings
| Property | Reversed-phased silica | Carbon |
| Surface functional groups | n-Alkyl, alkylaryl, aryl, residual hydroxyis | Aromatic-type acidic and basic functional groups |
| Specific surface area | 100-200 m²/g | 5-1000 m²/g |
| Type and extent of particle porosity | Open pores 60-80% | Open and closed pores 30-60% |
| Mean pore size and pore size distribution | 5-25 nm homogeneous | 1-1000 nm heterogeneous |
| Digits being effective to its measurement resolution |
A defined procedures, that can be assessed by anybody, for experiments and measurements.
|
Clinical analysis (臨床分析)The improvement of sensitivity and specificity on chromatography
[ultraspure substances]__[simple introduction] |
Target (Chait & Ebersole 1981) |
[ Markers on litter decomposition ]
Protocol for the quantificationExtraction of total lipids
5 g soil (powderization, if required)
In the test tube |
↓ + 5 ml chloroform Separation of PLFAs from total lipids
chloroform fraction (= total lipid) Identification and quantification of PFLAsIf molecular species can not be identified, the species is determined by a gas chromatograph mass spectrometer (so-called GC mass) |
PreparationPlate: silica gel wed. 0.25 mm (5 × 20 cm) MerkMaterials: purified lipid standard substance: GL (Cardiolypin), PG (Phosphatidyl glycerol), PE (Phosphatidyl ethanolamine) Developing liquid
1) chloroform:methanol:water = 65:25:5 ManipulationSpreadingmake a small pot (φ = 20-80 mm) of 60-100 ml sample at 1.5 cm from the bottom of plate by a capillary tube place 60-100 m solvent for 1-2 hr in a developing cabinet (to equilibrate the inisde) Start development prop the plate in a developing cabinet and leave them for 1.5-2 hours → when the developing sample reaches to the 2-3 cm from the top, the developing is terminated remove the plate from the cabinet and the solvent on the plate is diffused. Then, expose the plate to iodine →whenk spot is found out, mark it! (principle = iodine addition) Detection (I)
|
Detection (III). Ninhydrin reagent, used to detect amino acids
Cf. Extract component: phospholipids (90%), glycocide, fatty acids, etc. Examples on the results1st. PG was clearly distinguished. However, PG and CL were not clearly identified, due to the two spots of CL standard. (BR = brown-red)Sample Line Spot Length Rf Dittmer Nin. Stand. 17.1 PE 6.95 0.406 blue BR
PG 4.45 0.260 blue blue
CL-x1 4.75 0.278 blue blue
CL-x2 3.25 0.190 yellow yellow
20C left 16.95 a1 7.00 0.413 blue BR
a2 4.25 0.251 blue blue
a3 3.475 0.205 blue blue
20C right 16.95 b1 6.60 0.389 blue BR
b2 3.95 0.233 blue blue
b3 3.25 0.192 blue blue
37C 16.95 c1 6.70 0.395 blue BR
c2 4.375 0.258 blue blue
c3 3.45 0.204 blue blue
2nd The separations are insufficient. In particular, there were a few unclear spots made by ninhydrin reaction Sample Stand. 20C left 20C right 37C Line 16.4 16.55
Spot PE PG CL d1 d2 e1 e2 f1 f2
Length 4.075 3.35 4.05 4.05 3.2 3.9 3.05 3.95 3.05
Rf 0.248 0.202 0.247 0.245 0.193 0.236 0.184 0.239 0.184
Dittmer blue blue blue blue blue blue blue blue blue
Nin. - - - - - - - - -
|
Centrifugation (遠心分離)= centrifugal separationPrinciple: separation by using density difference - not by the mass i) centrifugal force, fr fr = mrω2
m: particle mass (kg)
G: relative centrifugal force, RCF = mrω2/mg = rω2/g Centrifugal (遠心分離器)= centrifugal machine, centrifugal precipitator, centrifugal separator, centrifuge, centrifuge machine or centrifuge separatorSwing-out roter 104/min_______Fixed angled roter: 2 × 105/min
Pellet (precipitate): hard-packed concentration of particles after centrifugation Supernatant: clarified liquid above the precipitate |
Separation of ERhomogenatet cells┃ centrigugate with 800-1000 × g, 5-10' + 0.2-0.4 M sucrose ┣━━━━━━━━━━━━━━━━┓_____⇑ not break organella ppt (nuclei, chloroplast)_____supernatant ┏━━━━━━━━━━━━━━━━┫ 10000-15000 × g, 15-20' ppt (mitochondria)_________supernatant = post-mitochondrial fraction ┏━━━━━━━━━━━━━━━━┫ 24000-40000 × g, 20-30' ppt (r-ER + microsome)_____supernatant ┏━━━━━━━━━━━━━━━━┫ ultracentrifuge with ┃━━━━━━━━━━━━━━━━┃ 102000-105000 × g, 2 hr ppt (free ribosomes polysomes) supernatant = S100 fraction Furthermore, ribosomes are separated from r-ER by the separation the addtion of deoxycholate-N (DOC), Nonidet P40 (NP40), polyethylene cethylethen (Bry 58) or Triton-X100 that uncouples lipids in r-ER *: g vs rpm = revolution per minutes S: unit of centrifugal separation |
|
Def. Isoelectric point (等電点), pI: 0 charge = net positive charge
Application: Purification and assay of enzymes (酵素の精製と分析)D-Glycerate dehydrogenese purified from spinach leaves (EC 1.1.1.29 D-Glycerate NAD oxidoreductase)COOH-CHOH-CH2OH (D-glycerate) + NAD+ ⇔ COOH-CO-CH2OH (Hydroxypyruvate) + NADH + H+ AimsFigure out enzyme purification techniques and enzyme characteristics.I. Purification procedureFresh spinach (Spinacia oleracea L.) leaves (ca. 1 kg)
↓ 0.001 M 1-litter EDTA (pH 8.0)
Add 13.3 g of AmSO4/100 ml of sup. with gentle
Adjust pH at 4.85 with addition of 1 N acetic acid 10-20 h under cold (4°C)
Add 12.1 g of AmSO4/100 ml of sup. with gentle stirring
Dialyze the sample against 5 l of the same buffer for 3 hr with 2 changes of buffer
Stir slowly for 15 min.
Add 25 g of AmSO4/100 ml of sup. |
↓ - Fraction VII
End Table 1. Protein concentration in each fraction. C = concentration
Fraction# Dilution 1st 2nd 3rd C (mg/ml)
I × 100 0.520 0.538 0.533 9.533
II × 200 0.485 0.455 0.470 16.53
III × 100 0.281 0.299 0.290 4.733
IV × 40 0.304 0.320 0.312 2.052
V × 10 0.380 0.390 0.385 0.660
VI × 10 0.200 0.208 0.204 0.307
VII × 100 0.210 0.208 0.209 3.133
II. Enzyme assayLy-hydroxypruvate 0.008 M (pH 6.0) 0.25 ml NADH 1 mg/ml 0.2 ml Ammonium sulfate 0.4 M 0.5 ml Phosphate buffer 0.6 M (pH 6.0) 1.0 ml Enzyme (sample) 0.1 ml Deionized water Final volume 3.1 mlEnzyme dilution buffer: 0.01 M phosphate buffer (pH 6.0) Results (Table 1) I. Protein concentration in each fraction II. Enzyme activity When a sample is too dense to measure the concentration, re-measurement should be conducted after the dilution. Measure NADH-NAD+ activity. Ex. NAD oxydase: NADH + O2 - NAD+ in 1 ml reactive solution (Table 2) Measurement of Km Fraction #VII was diluted to 1/2, 1/4, 1/8, 1/10, and 1/16, using 0.008 M substrates. Thereafter, measure the activities by the enzyme assay described above. (Results, omitted) Thermal inactivation (熱失活) Methods: Treat Fraction VII at 40°C for 10, 20, 40 and 80 min, and at 50°C for 1, 2, 3, 4, and 5 min. Measure the activities by enzyme assay method described above Effect of salt on enzyme activity (塩の効果) Methods: Measure reaction pace by various salts, i.e., KCl and NaCl, (0.4 M) with different soaking periods (0, 30, 60, 90, 120, 180, and 210 min., if possible soak longer time) and cmpare with (NH4)2SO4 Table 2. Results. SA = specific activity (units/ml·protein) Frac- Volume Protein Activity SA Purifi- Yield tion# (ml) (mg/ml) (unit/ml) cation (%) I 232 9.533 5.114 0.5365 1.0 100.0 II 50 16.53 19.27 1.1658 2.17 81.2 III 45.5 4.733 18.625 3.9351 7.33 71.4 IV 17 2.052 14.6 7.1150 13.26 20.9 V 34 0.660 3.515 5.3258 9.93 10.1 VI ? 0.303 1.515 4.9349 9.20 ? VII 3 3.133 17.7 5.6495 10.53 4.5 |
Nitrogen1. Quantitative analysis of nitrogenUV absorbance = adjust at 280 nm Absorbance Ex. Aromatic amino acid: phenylalanine: 250 nm = 190 mol Ab., Tryptophane: 278 nm = 5500 mol Ab., Trypsin: 275 nm = 1340 mol Ab. 2. Turbid method: acidic - low solubilitySample: protein 0.5-1.5 mg/ml + 1.25% TCA (Trichloroacetic acid) 4 ml 3. Lowry's method: Phenol reagent
A: 2% Na2CO3 with 0.1N NaOH D and E should be prepared just before the using (within 24 hr), because those are unstable solution. F: phenol reagent 1N |
Procedure
sample (5-100 g protein) 0.6 ml Dye |
|
(Walter 1983) Dry reagent chemistries in clinical analysisDry reagent chemistryprogress of quantitative measurments based on analytical chemistry on iatrochemistry19c: invented litmus paper 1970s': developed dry reagent chemistry for measuring blood elements quantitatively
__Reagent zone__ ↔ Large amount of samples by several steps on many medical examinations Components in dry reagent carrier
types of chemical reaction monitored |
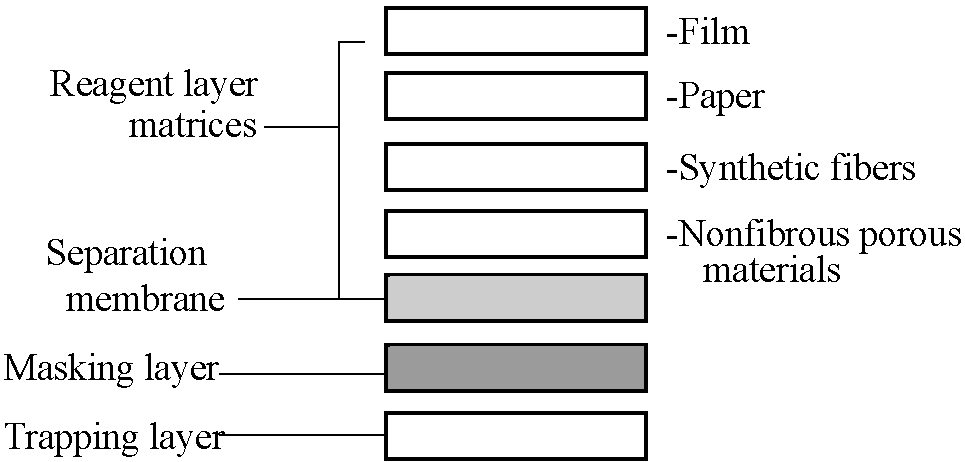 Fig. 1. Basic components of dry reagent chemistry carriers. |
|
(Abbreviations: RIA = radioimmunoassay, FIA = fluorescent immunoassay, ELISA = enzyme-linked immuoassay, EMIT = enzyme-multiplied immunoassay) Table 1. Immunoassay comparison
RIA FLA EIA
Sensitivity High (ng-pg) High (ng-pg) High (ng-pg)
Specificity High High High
Speed Days Days Hours
Reagents Short shelf Reasonable Long shelf
life shelf life life
Equipment Scintillation or UV monitor Spectro-
required gamma counter photometer
Personnel Skilled with Skilled Minimal
license training
Cost $7/test $5/test $2/test
Table 2. EIA comparison
ELISA ↔ EMIT |
* hapten (ハプテン, 付着体): small molecules that elicit an immune response only when attached to a large carrier such as a protein Ex. urushiol
__sample____________buffer/reagents__ Polystyrene tube coated with antigen ← Incubate at 37°C ← Wash 3X ← Serum added ← Incubate at 37°C ← Wash 3X ← Enzyme-labeled antiserum added ← Incubate at 37°C ← Wash 3X ← Substrate added ← Visible color change ← Spectrophotometer absorption readings Fig. 6. ELISA steps for serodiagnosis of diseases |
Medium for water culture (hydroponics). Observation points: pH, leaf colors, and dry weight
|
International System of Units, SI (国際単位系)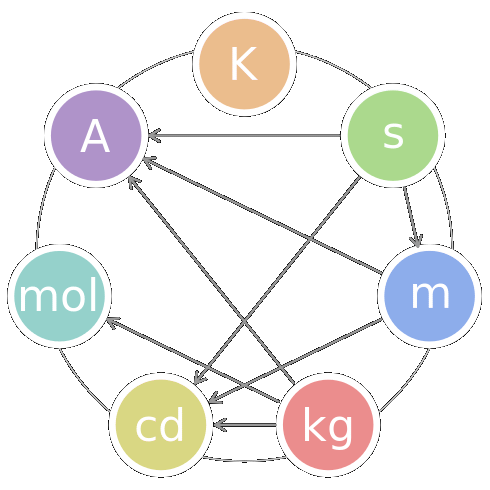 The seven SI base units
The seven SI base unitskelvin (temperature) second (time) meter (length) kilogram (mass) candela (luminous intensity) mole (amount of substance) ampere (electric current) Metric system (メートル法)1 mm = 1/1000 m1 μm = 1/1000 mm 1 nm = 1/1000 μm 1 Å = 1/10 nm |
Cardinal numbers (基数)100:____________ one hundred101:____________ one hundred (and) one 1,000:___________one thousand 100,000:_________one hundred thousand 30,000,000,000:___three followed by ten zero 1,000,000:_______ one million 1,000,000,000:____one billion 1,000,000,000,000: one trillion 1015:____________one quadrillion 1018:____________one quintillion 1021:____________one sextillion 1024:____________one septillion 1027:____________one octillion 1030:____________one nonillion 1033:____________one decillion 1010: one followed by ten zeros (ten to the tenth) RatioPercent vs percentagePercent is a number."20 percent (20%) of people have pets". You cannot say "20 percentage of people have pets". Percentage means a part of the whole."a small percentage of people have pets". Per mille1‰ = 10-3 = 0.1% (⇒ isotope) |
An act, process, or instance of validating; especially : the determination of the degree of validity of a measuring device (Merrian-Webster Online)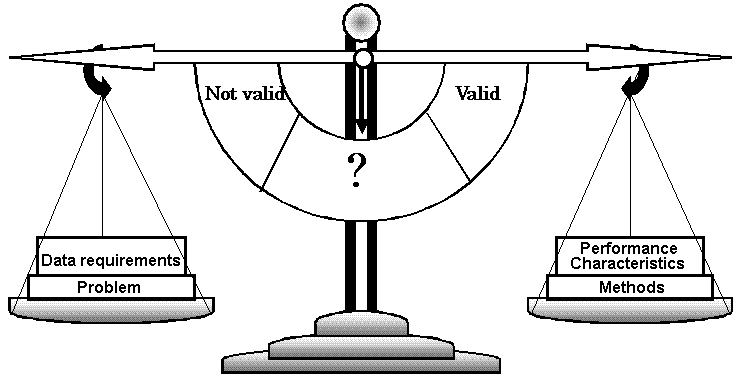 Fig. Basic concept of the validation process like a balance. Importance of validationTechnique: scientific principle useful for providing compositional informationSpectrophotometry Procedure: Distinct adaptation of a technique for a selected measurement purposePararosaniline method for measurement of sulfur dioxide |
Method: Written directions necessary to use a method
ASTM D2914 - Standard Test Method for the Sulfur Dioxide Content of the Atmosphere (West Gaeke Method) Protocol: Set of definitive directions that must be followed, without exception, if the analytical results are to be accepted for a given purposeEPA Reference Method for the Determination of Sulfur Dioxide in the Atmosphere (Pararosaniline Method)  Fig. Collaborative test process. 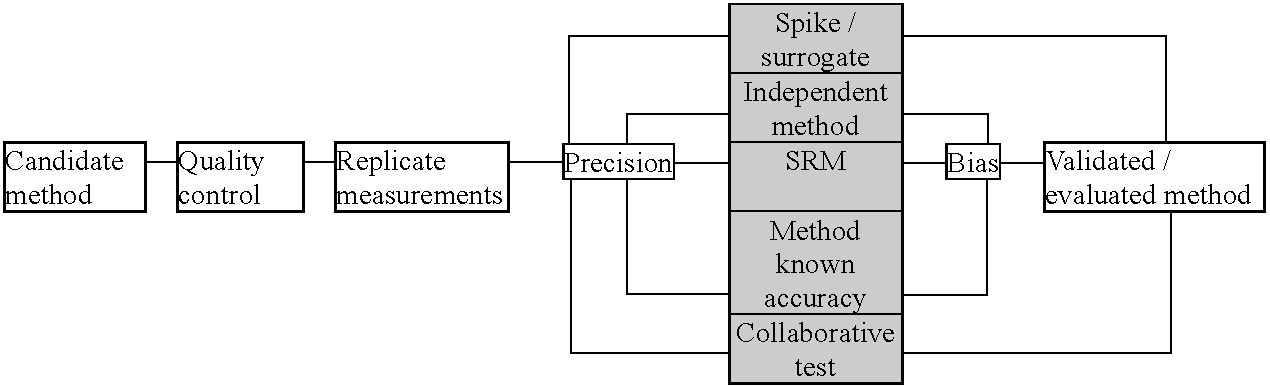 Fig. 3. General process for evaluation/validation of methodology. |