(Upload on March 3 2020) [ 日本語 | English ]
Mount Usu / Sarobetsu post-mined peatland
From left: Crater basin in 1986 and 2006. Cottongrass / Daylily
HOME > Lecture catalog / Research summary > Glossary > Biology > Cell biology > Gene
Gene (遺伝子)sequence of nucleotides in DNA or RNA that encodes the synthesis of a gene product, either RNA or proteinSynthetic biology (合成生物学) |
|
|
1869 Miescher, Friedrich (1844-1895): nuclein extracted from pus 1871 Hoppe-Seyler EFI (1825-1895) & Miescher F nuclein extracted from red blood cell, yeast, yolk, salmon soft roe, etc → they did not detect the functionStructurea) nucleotide = [base]–[sugar]–[phosphate acid]
N-base___ nucleoside cf. nucleoside: [base]–[sugar] → triphosphate deoxynucleoside, distributed in nuclei
A-dR: deoxyadenosine_A-R: adenosine___dATP
DNA (= deoxyribose): H bounded at C2' ↔ RNA (= ribose): OH bounded at C2'
relative base lg (μm) MW
volume pair
DNA virus SV40 4,000 1.6 3.1·106
λ phage 53,000 16 31·106
bacteria E. coli 3,400,000 12,000 2,320·106
RNA m-RNA 5% random -4·105
t-RNA 15% 70-80 2.5·104
r-RNA 80% 120 3.6·104
1,700 0.55·106
3,700 1.2·106
Purine group (プリン族): purine nuclei (consisting of five-membered and six-membered rings) = adenine (A) and guanine (G) → C1 on the pentose replaced from H on N9 when nucleic acid is formed
|
Pyrimidine group (ピリミジン族): Pyrimidine nuculei (consisting of six-membered rings) = Cytosine (C), Uracil (U), Thymine (T) → C1 on the pentose replaced from H on N3 when nucleic acid is formed
Abnormal base (異常塩基)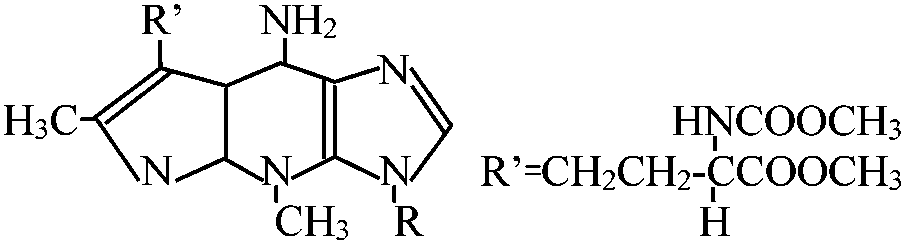 R = base Y R = base Y2'O-methyl nucleoside [2'Om (A, C, G, or U)] 7-Methyl guanosine (7mG)___Inosine (I) 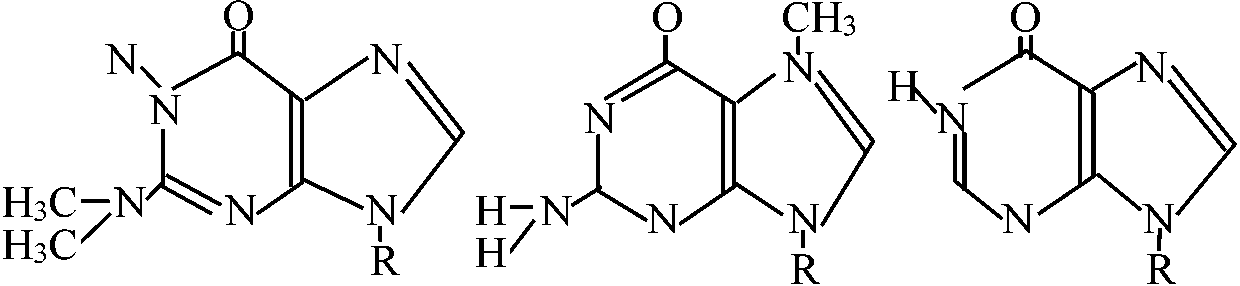 N2-Dimethyl guanosine (2dmG) N6-ΔIsopentenyl 2 methylthioadenosine (2ms6iA) 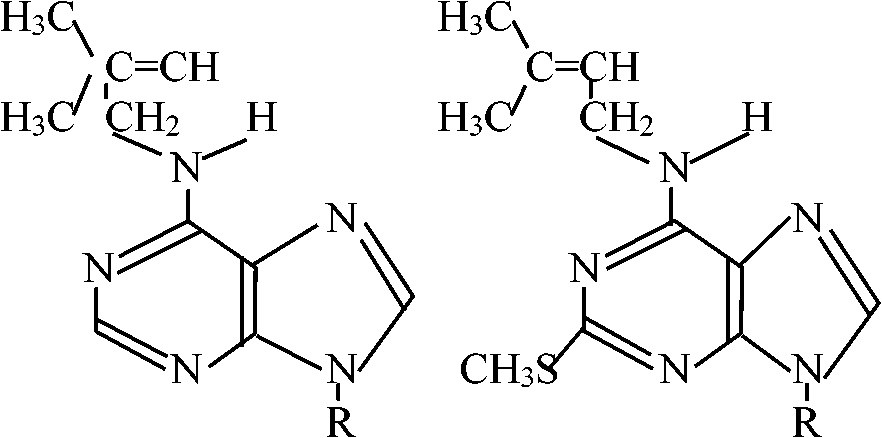 N6-ΔIsopentenyl adenosine 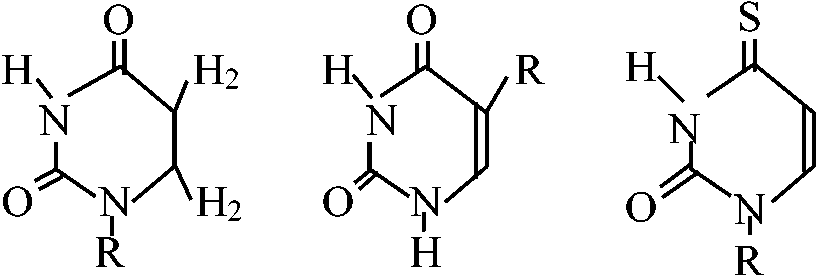 Dihydrouridine (D) Pseudouridine (ψ)_4-Thiouridine (4SU) |
Extraction of DNA from DNA-protein complexDNA-protein complex (including RNA)
0.1M Tris-HCl (pH 7.5-8.0), 0.1-0.2 M NaCl, 1-5 mM EDTA, homogenized with 1-2% SDS |
|
Circle DNA (環状DNA) i) Open circle DNA (o.c. DNA) ii) Closed circular (c.c. DNA) (= coralently closed circular DNA, superhelix, supertwist, super coil)
relaxed circlar DNA____super coil |
DNA polymerase (DNA合成酵素)1956 Kornberg et al.: discovered from E.coli in the experiment of in vitro DNA replication
dXTP = uptake into primer DNA
MW 10900, single-strand peptide, one SS-bond
response to both 5'-end and 3'-end
5' → 3': transfer the next nucleotide to 3'-OH, indicator free 3'-OH
discovered defective mutant of DNA polymerase I(A) from 3478 strains → DNA polymerase I is not essential for DNA synthesis 1969 Cairns et al. +1970 Kornberg & Gefter + 1970 Moses & Richardson + 1970 Knippers DNA polymerase II 1970 Kornberg TDNA polymerase III: RNA as primer, DNA as template, 5' → 3' 1970 Temin, Baltimorediscovered reverse transcriptase = RNA-dependent DNA polymerase 1971 Wang: E. coli W-protein – DNA binding protein
dna genge: engaged in gene replication DNA polymeraseE. coli: poly I(109Kd), II(120Kd), III(140Kd) & III*Mammals: poly I(α), II(β), III(γ) DNA polymerase IDNA dependentOkazaki fragment lacks the primer RNA polymerase does not need the primer → A hypothesis proposed by Okazaki is that: firstly RNA is formed and then DNA is formed by removing RNA later DNA polymerase II
DNA polymerase III* |α 140| Poly III (dna E, pol C)
|ε 25 |
|θ 10 |
|J 83
|γ 32 dna Z |factor II
|δ 32 factor III |
|β 40 factor I – copol III*
Three DNA polymerases, α, β and γ, in eukaryotesα: 12-13 MW
high ion intensity > 25 mM
utilizing oligo12-18, poly(rA)
favor oligo |
Plant DNA polymerase1974 Mory, Chew & Sarid: DNA polymerase extracted from wheat1978 Stevens & Bryant mashing up pea seedlings &larr ; collecting supernatant after centrifugation → ammonium sulfate concentration gradient → extracting animal-like α, β DNA plymerases 1979 Horemann & Follmann
DNA synthesis by Vicia fava DNA repair (DNA修復)1) Gilbert
neck____Enzyme (= ligase joining enzyme) 3) 1967 Gilbert
Trigger of DNA synthesis (DNA合生開始)Semi-conservative DNA replication (半保存的DNA複製)1960 Matsuoka: Chlamydomonus (eukaryotes), 1961 Simon: Hela cellTable. Structure of polynucleotide double helices 1967-1970 Inman
λ-DNA: eye shape = double strand
NA, cytokinine c–basic + 2M ammonium acetate 
30, 32, 41, 42, 43, 45, 46, 47, 56, 62 Replication (複製)1) Eukaryote
nucleolus-DNA, r-RNA genes → replications [special example] 1st: Caris type (θ) → 2nd: Rolling type 2’) Mitochondria DNA (Robberson, Kasamatsu & Vinograd 1972)
a___________b_____________c Sites for DNA replication (DNA複製の場所)Hypothesis: eukaryotic body cells (2n → DNA replication starting for nuclear membrane1965 Sparvoli, Gay & Kaufmann Tradescantia - prochromosome 1965 DuPrawchromatin fiber ≈ 230Å 1970 Comings & Okadaat metaphase on Chinese hamster, etc. → chromatin bounded with nuclear membreane 1973 Huberman, Tsai & Deich1976 Sparvoli, Galli, Mossa, & Paris
Haplopappus gracilis, n = 2 |
| Sample | Relative humidity (%) | Residues per turn | Translation per residue (Å) | Angle between base planes and helix axis | Dihedral angle between base planes | Sugar conformation |
| DNA-A, Na+ | 75 | 11 | 2.55 | 70o | 16° | C3'-endo |
| DNA-B, Na+ | 95 | 10 | 3.46 | |||
| DNA-B, Li+ | 66 | 10 | 3.37 | 88° | 5° | C2'-endo |
| DNA-C, Li+ | 66 | 9.3 | 3.32 | 84° | 10° | C2'-endo |
| DNA-RNA hybrid, Na+ | 75 | 11 | 2.62 | 70° | ||
| Yeast RNA fragments | 75 | 10 or 11 | 2.9 or 2.64 | |||
| Reovirus α or β forms | 75 | 10 or 11 | 3.0 or 2.73 | |||
| poly rA·rU | 10 or 11 | 3.1 | ||||
| poly rI·rC | 12 | 3.0 | ||||
| poly rCH+·rC* | 12 | 3.11 | ||||
| poly rAH+·rAH+* | 8 | 3.8 |
|
Alkaptonuria or alcaptonuria アルカプトン症 Garrod Archibald (1857-1936, England) 1902 "The incidence of alkaptonuria: a study in chemical individuality" 1923 "Inborn errors of metabolism (先天性代謝異常)" Alkaptonuria, cystinuria (シスチン尿症), pentosuria (ペントース尿症), achromia or albinism (色素欠乏症) One gene-one enzyme theoryBeadle, George Wells (1903-1989)Tatum, Edward Lawrie (1909-1975) 1941 Beadle & Tatum: proposed one gene-one enzyme theory |
= A + G + C + U
|
16S ribosomal RNA (16S rRNA)Prokaryota - 16S rRNA geneused in reconstructing phylogenies, because of the slow evolution rate of this region of the gene 16S ribosomal databasesEzBioCloud (former EzTaxon)Ribosomal Database Project (RDP) SILVA GreenGenes Gut bacteria: global rumen census datasets (Henderson et al. 2019) |
Reversed genetics (改変遺伝学)gene engineering (遺伝子工学) = gene manipulation (遺伝子操作)
isolation of gene → assay of gene structure → restriction map (制限酵素地図) designed alteration of the gene gene structure can be altered freely by the researchers who use reversed genetics
fraction test of the mutant (mutated) genes (J Genetics 57: 581-598, 1982) |
I. identification of signal DNA sequencefinding out the components responsible for the function→ fractionation of extract → purification → rreconstitution → addition of regulatory components → establishment of basic transcription system: the amount of transcription can be adjusted by using different developmental stages → clarifying the components of regulation II. types of cellsBasic transcriptive function=============== DNA signal: utilized heterogenous cells Regulatory function -------------------------- cell that expressed, per se ____________________________↓ when cells containing different genes are inserted, endogenous or exogenous genes can be detected  III. fertilized egg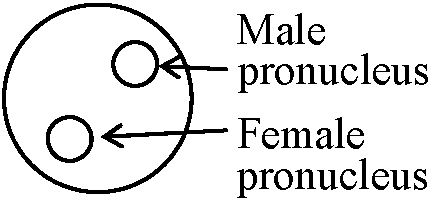 mouse/Drosophila
mouse/Drosophila
→ test of regulated expression → identification of signal DNA sequences Finding out the component responsible for the regulated function |
|
(Suzuki & Brown 1972) Isolation of sil fibroin mRNAcrude mRNA for globin mRNA (only the way at that time) (Williamson & Paul 1972)crude mRNA: labeling → hybridization with DNA → -104 genes/haploid: required pure DNA Table. Bombyx mori: the amount of amino acids is measurable, and the sequence is partially determined Glycine Alanie Serine Tyrosine Others mol% 45 29 12 5 9 genetic codon GGX GCY UCZ or AG[U/G] UA[U/C]
G/G content = high
60% (w/w):_____(Gly-Ala-Gly-Ala-Gly-[Ser-Gly-(Ala-Gly)2]8- in Rnase T1 (cut 3'-side of G) Finger print: consider UCZ case (many Z = A)
Residue G XG CYG XUCZG Total Profile made by UD260, 32P
Advantages of silk fibroin = 16000 nucleotides, 4.4% of total DNA Detection of fibroin genes1972 Suzuki, Gage & Brown
Ex. fibroin mRNA 90%: 1 copy/haploid → 90 not matched with the fact
DNA in rear-part silk gland cell ........ 1-3
fibroin gene = 0.002 × 2 (%) of total genome
4 kinds out of ca. 50-70% kinds Structural analysis of the genesIntron discovery (イントロン) (Cold Spring Harbor Symposium, Quantitative Biology 1977)Intron was firstly confirmed by using adeno-virus 2 DNA (chromatin)
├──→
──┼───────── early gene
5' ──┼───────── late gene (functional)
│←──
DNA-RNA hybridization: EM level
Mature mRNA ========== "intron" sequence all sequences are transcribed at first, and then unnecessary portions are discarded complementary DNA obtained from the mRNA of mouse globrin (Tilghman et al. 1977) R-loop method (Thomas et al. 1976)a)__ 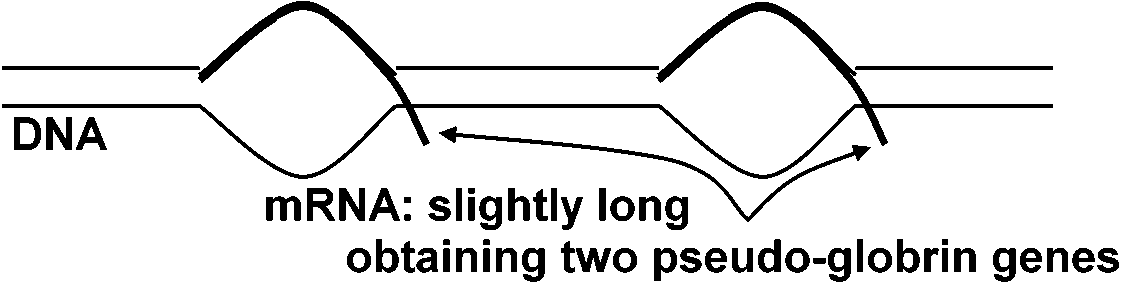
5' 3'
────┼────────────┼───
────┼────────────┼───
└──↑─────────┘
1 kb 1kb intron 15 kb ⇒ mRNA = 16 kb
the lengths of fibloin fibers are different between the strains - the lengths of DNA are proportionally changed b) intron existing like this in nature
── exon, ━━ itron
⇒ 1 globrin gene(Tsujimoto & Suzuki 1979ab) ⇒ (ProNAS 74: 4406, 1980)
========================⇒ mRNA 
Southern blottingTotal DNA↓ restriction enzyme A DNA digest ↓ electrophoresis ↓ EtBr 
 0 × ssc
0 × ssc20 × 0.15 M NaCl, 0.015 M sodium citrate radioautograph after single strand made by alkaline treatment
⇒ two fragments obtained by using enzyme A 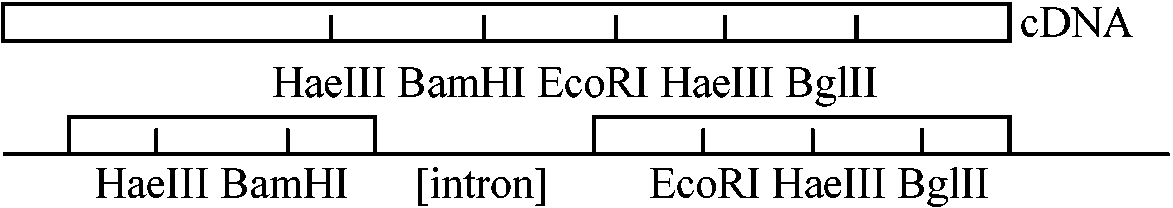 1978 Tilghman et al. 
friend cells 8 l |
R-loop
β+ thalassemia (地中海性貧血症) – produced globrin somehow ×: mutation having the genes in the outside
mutation [normal = G, β+ = A] at 21 bp
ex. 1______int. 1_____ex. 2 (Kole & Weissman 1982) in vivo splicingHela cell
_________________Nomal splicing__Abnormal splicing
Hela cell whole extract
Trancation assay Synthesizing precursor in vitro
↓ phenol extraction ↓ no phenol
↓ new + extract ↓ extract
↓ incubation ↓ incubation
↓ ↓
RNA degraded Stable
probably proteins connected to the synthesized RNA - (possibly) due to splicing
Cell → Total cellular RNA: *32P
SI mapping method: method to determine initiation point actual signal for starting transcription (ex. AGT) = promoter Hybrids arrest = ex. clone for few numbers of mRNA Use of in vitro transcription system (生体外翻訳系利用)RNA polymerase of eukaryotes
Type I gene = rRNA: rDNA = class I gene firstly, transcription of type III gene was analyzed Type I gene: ETS (external transcribed sequence), ITS (internal transcribed sequence) and coding regionType II gene: modified RNA polymerase II genes
Manley et al. (1980): Hela cell extract ------------===============▆▆▆▆▆▆▆▆▆▆▆ =========▶ ----------------===========▆▆▆▆▆▆▆▆▆▆▆ ---------------------======▆▆▆▆▆▆▆▆▆▆▆ -------------------------==▆▆▆▆▆▆▆▆▆▆▆
_________↑__↑____↑__ ↑ these can be separated by an electrophoresis because the lengths (or masses) of mRNA are different
signal can be activated when the region of -29~+6 is present
Used the method proposed by Manley et al. = 0.4 M (NH4)2SO4 |
|
1952 Mitchell MB & Mitchell HK: Neurospora crassa (red bread mold)
slow-growing mutant, called poky
poky (mother, ♀) → poky (all children) → mutation occurred on mitochondria 1981 Anderson et al.: the sequence of mitochondrial DNA of human16569 base pairs - circular gene Mitochondrial gene (mt gene, ミトコンドリア遺伝子)circular DNA (genome size = DNA size, kbp = kilo base pair)unstable: 16kbp (mammals). 20-100 kbp (protozoa, fungi), 60 kbp (tobacco), 2400 kbp (muskmelon) |
Chloroplast gene (葉緑体遺伝子, chl gene)1888 Schimper FW: chloroplants - autonomous growth → endosymbiosis1909 Correns C: non-Mendelian inheritance - proposed plasma gene 1959 Stocking CR: chloroplasts - 3H-thymidine uptake → DNA 1960 Iwamura T (岩村達一): DNA extracted from chloroplasts 1962 Ris H: DNA fibers within chloroplants, observed by EM 1963 Sager R & Ishida M (石田正弘): chlamydomoas DNA extracted from chloroplats (later confirmed DNA in all plastids) Chloroplast genomedouble-stranded circular DNA1980 Koessal: Euglena. 1980 Takaiwa & Sugiura: tobbaco
Determined the sequence of chloroplast DNA (*: tRNA) Plasmid (プラスミド) |
Assay by using cultured cells (培養細胞利用検定法)interferon (human) β gene in plasmid (PSV2-Ecogpt)↓ CalPh gel ↓ mouse FM3A 2.3 copies of β genes/cell
New castle desease virus = poly(I):poly(c) (Ohno & Taniguchi 1982)
Chitin lysozyme gene11 |
↓ (culture: various parts collected from chitin) 1: + steroid hormone →
cells endocytose fluorescent substance to thymidine kinase Δ-161~+15: thimidine kinase gene Assay system by using fertilized eggBrinster et al. (1982)
tK molecules -cd +cd 0 8.5 8.8 20 8.3 12.2 200 25.3 270 2000 190 2400 -1.7 kbp = remaining: 0K, -99 bp leaving: 0K |
DNA marker (DNAマーカー)RAMP (random amplified microsatellite polymorphism)SSCP (single strand conformational polymorphism) PCR-RFLP [CAPS (cleaved amplified polymorphic sequences)] AFLP (amplified fragment length polymorphism) Terminal Restriction Fragment Length Polymorphism (T-RFLP) ProtocolPreparing a sample for T-RFLP analysis is a two step process. First, PCR amplify the desired region of DNA using a HEX labeled forward primer and/or a FAM labeled reverse primer. Second, digest the PCR product with one or more restriction enzymes. The details of this process follow.PCR amplify DNA using labeled primer(s)Working concentrations of components:
Component = Working concentration
Reaction
Component 25 μL 50 μL 75 μL 100 μL
Forward primer 0.50 1.00 1.50 2.00
HEX forward pr.* 0.75 1.50 2.25 3.00
Reverse primer 0.50 1.00 1.50 2.00
FAM reverse pr.* 0.75 1.50 2.25 3.00
Buffer 2.50 5.00 7.50 10.00
MgCl2 3.75 7.50 11.25 15.00
dNTP's 2.50 5.00 7.50 10.00
BSA 0.05 0.10 0.15 0.20
milliQ H2O 13.35 26.70 40.05 53.40
Taq 0.10 0.20 0.30 0.40
Template 1.50 3.00 4.50 6.00
* NOTE: Run separate PCR reactions for each labeled primer (i.e., run HEX labeled forward primer with unlabeled reverse primer; run FAM labeled reverse primer with unlabeled forward primer). |
Thermal cycler (サーマルサイクラー)= thermocycler, PCR machine or DNA amplifiera laboratory apparatus most commonly used to amplify segments of DNA via the PCR 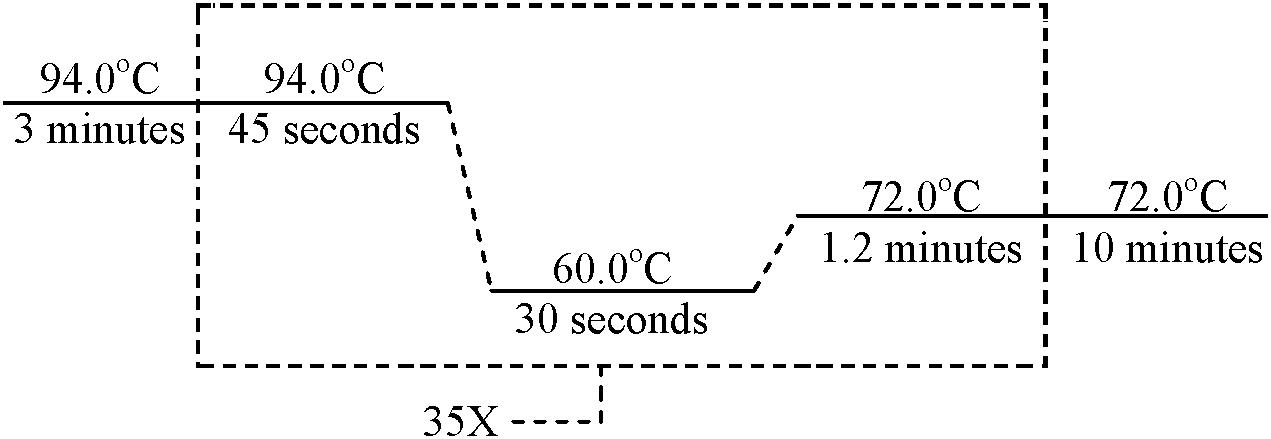 Using the parameters above, amplify DNA using labeled primer(s) Check PCR product by agarose gel electrophoresis
- If positive, purify product Digest the PCR productWorking concentrations of components:Component Working concentration Labeled PCR product* 250-300 ng (each) per digest GibcoBRL REact buffer 10X (restriction enzyme specific) GibcoBRL restriction enzyme 10 U/μL milliQ H2O na * NOTE: If you have both HEX and FAM labeled product, combine 250–300 ng of each labeled product in a single digest (i.e., 250 – 300 ng of HEX labeled product combined with 250–300 ng of FAM labeled product to give a total of 500–600 ng of labeled product in a 20 μL digest) Volumes (μL) of components per digest using working concentrations noted above:
Component Volume (20 μL digest)
Labeled PCR product depends on PCR product
concentration
Buffer 2.00
Restriction enzyme 1.50
milliQ H2O dilute digest to 20 μL
(PCR product + H2O = 16.5 μL
+ buff. + re. enz. = 20 μL)
Digest purified PCR product with preferred restriction enzyme(s) - If digesting with multiple restriction enzymes, do a separate digest for each restriction enzyme Digested product is ready for T-RFLP analysis Protocol provided by Brian Wade at the MSU Center for Microbial Ecology |
|
multidisciplinary science focusing on living systems and organisms
including designing and constructing biological modules, biological systems, and biological machines 1910 Leduc, Stéphane: firstly used "synthetic biology"2003 Knight, Tom: BioBrick plasmids → iGEM 2010 Gibson DG, et al.: synthetic bacterial genome Mycoplasma mycoides JCVI-syn1.0 ☛ bioethics |